PhD student Turmagambetova
A., prof. Ivashchenko A.
al-Farabi Kazakh National
University, Almaty, Kazakhstan
HYDROPATHY OF EXONS, INTRONS AND SPLICING SITES
IN HUMAN ONCOGENES
Due to computer methods there is an opportunity to
find out and to predict many biochemical and molecular-genetic properties of
nucleotide DNA sequences. The interest to study of genes exon-intron
organization has increased in connection with data accession about new
properties and functions of introns /1/. The presence of regulating sites of
genes expression in introns was shown. The introns role in increase of a
variety of products of genes expression by alternative splicing was
demonstrated. Fairly often wrong splicing results in pathologies, which
represents the large interest from the practical point of view. For example,
establishment hydropathy of splice sites and prediction of alternative splicing
for genes responsible for gastrointestinal tract cancer development is
important.
Methods
Nucleotide sequence of genes was extracted from GenBank (http://www.ncbi.nlm.nih.gov/).
Prediction of alternative splicing is based on exon-intron hydropathy
determination. In order to distinguish biochemical properties of the pre-mRNA
of genes responsible for gastrointestinal tract cancer we define a general
hydropathy profile as described below. A hydropathy coefficient HC(N) may be
associated with each base N. Coefficients provided in /2/, are HC(A) = -1.07,
HC(C) = -0.76, HC(G) = -1.36 and HC(U) = -0.76. We compute an average
hydropathy value for each exon and intron as follows. For each base, its number
of occurrences in the exon/intron is multiplied by its hydropathy coefficients.
Summing over all the bases yields the average hydropathy value.
We used the nucleotides in positions -8 to +30 at the 5’ splice sites
and nucleotides in positions -35 to +8 at the 3’ splice sites to build
hydropathy profile of the splice sites of different intron types. For each
intron set and each position an average hydropathy value is computed.
Hydropathy coefficients are associated to each base. Average hydropathy value
has counted by multiplying each coefficient by the occurrence at the sets at
the given position.
Results
We have determined that hydropathy of exon and intron depends on their
nucleotide composition. Usually exons are more hydrophilic, than introns. Thus,
introns mostly form a pre-mRNA core while
exons are mainly located on surface of molecule. Example
of several genes responsible for gastrointestinal tract cancer development
shows the difference of exons and introns hydropathy, for genes without
alternative splicing and having some alternative products. It was established
that in genes without alternative splicing hydropathy of exons lower, than that
of introns. It is depicted in Figure 1 that exons have negative hydropathy and
introns have positive one. The genes with alternative splicing have sites with
different hydropathy of exons and introns. For these genes there are sections
where hydropathy of exons and introns differs insignificantly (Figure 1). On
the basis of these data it is possible to assume, that places with an
insignificantly differ hydropathy of exons and introns are alternatively
spliced.
The protein encoded by CD44 gene is a cell-surface glycoprotein involved
in cell-cell interactions, cell adhesion and migration. It is a receptor for hyaluronic acid and can also interact with other ligands, such as osteopontin,
collagens, and matrix metalloproteinases. This
protein participates in a wide variety of cellular functions including
lymphocyte activation, recirculation and homing, hematopoiesis,
and tumor metastasis. 17 named isoforms of this
alternative splicing of CD44 gene and the majority of isoforms
may be related to tumor metastasis. In some cases
alternative splicing results in an alternative translation frame or premature termination of translation /3-5/. For
pre-mRNA of CD44 gene exons 3, 4, 5 and 17 are
alternatively spliced (Figure 1). Hydropathy of 6,
7, 8, 9, 10, 11, 13, 14 exons are precisely differs from hydropathy
of adjacent introns. These exons
consist of strongly hydrophilic extracellular domain. The choice of these
splice sites is carried out through special positive or negative trans-acting factors.
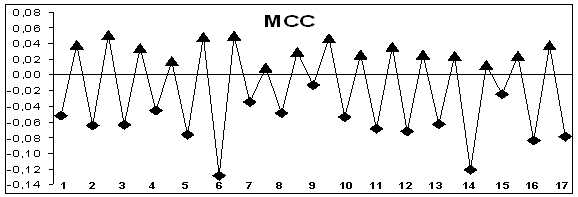
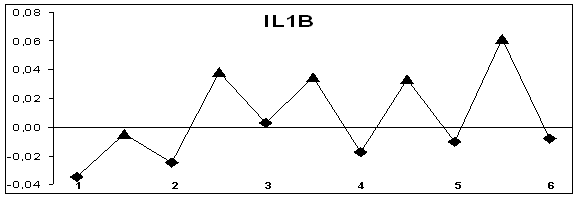
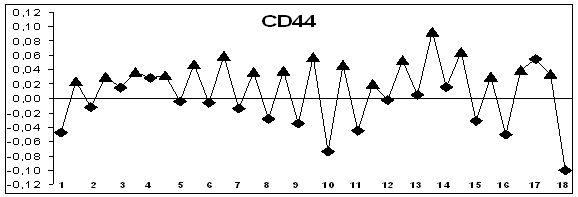
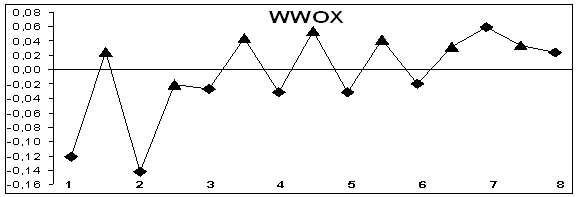
Figure 1. Hydropathy profiles of exons and introns of genes without
splicing (MCC, IL1B) and alternatively splicing genes (CD44, WWOX); õ– axis – exons (–♦– numbered) and introns (–▲–), ó-axis – base
hydropathy coefficients.
Negative
regulators would prevent the inclusion of variant exon sequences and thus give
rise to the expression of the standard form of CD44 (CD44s) in most tissues. Alternatively, choice of a specific
splice isoforms could be achieved by induced positive regulators that select
the corresponding variant exon combination. In tumor cells, altered splicing
patterns would then by caused by the up-regulation of such positive factors.
Gene WWOX
encodes a protein which contains 2 WW domains and a short dehydrogenase/reductase
domain (SRD). WW domain-containing proteins are found in all eukaryotes and
play an important role in the regulation of a wide variety of cellular functions
such as protein degradation, transcription, and RNA splicing. The highest
normal expression of this gene is detected in hormonally regulated tissues such
as testis, ovary, and prostate. This expression pattern and the presence of an
SRD domain suggest a role for this gene in steroid metabolism. The encoded
protein is more than 90% identical to the mouse protein, which is an essential
mediator of tumor necrosis factor-alpha-induced apoptosis, suggesting a
similar, important role in apoptosis for the human protein. In addition, there
is evidence that this gene behaves as a suppressor of tumor growth /6, 7/. It is depicted in Figure 1 that alternatively spliced only
3’ end of pre-mRNA (exons 6 and 7). According to previous works by others all
alternative variants of proteins differ only on 3' end. This study suggests
that it is possible to predict places of alternative splicing by given method. This
method is insufficiently if gene has special elements that regulate alternative
splicing.
Introns
are more hydrophobic, contact to each other and form core of pre-mRNA molecule. Mostly hydrophilic exons located predominantly on surface
of molecule as well as the 5' splice sites
and the 3' splice sites that facilitate
interaction and ligation of exons. Hence, it is
possible to assume, that the hydropathy profiles of the 5' splice sites and the 3'
splice sites should be similar. We define hydropathy profiles of all splice
sites of gene. We used the nucleotides in position -8 to +8 at the 5’ splice
sites and nucleotides in positions +8 to -8 at the 3' splice sites. The average
hydropathy value of all splice cites is computed to build hydropathy profile of
the splice sites of gene. Further the difference of hydropathy profiles of 5'
splice sites and 3' splice sites is shown by the example of several genes
responsible for gastrointestinal tract cancer development. We point out that
the 3' end of intron always more hydrophobic, than other parts of the 5' splice
sites and 3' splice sites.
Except for introns containing a conservative sequence
on 5' splice sites and 3' splice sites, there is a small group of introns with
other terminal dinucleotides: AT-AC introns. There are U12 snRNP
- dependent introns which are excised by spliceosomes containing U4atac,
U6atac, U5, U11 and U12 snRNPs. It is the minor class
of introns the quantity of the AT-AC introns on two orders is less, than usual introns. The molecular mechanism of splicing is based on
complementary binds and hydropathy interactions of pre-mRNA nucleotides and snRNA. These interactions play a significant role in
splicing catalysis. In order to distinguish biochemical properties of
the splice sites of genes responsible for gastrointestinal tract cancer we have
defined a general hydropathy profile as described in methods. There are at least two classes of splice sites on human
genome which differ in nucleotide consensus. There are two types of splice
sites for U2 (GT-AG and GC-AG) and U12 (GT-AG and AT-AC) snRNA.
Spliceosoma
apparatus is highly conservative that is why conservative regions must exist
near splice sites to provide signals for splice site recognition. Due to the
degeneracy of motives for U2 (U12)-type introns it is clearly that not always
nature of spliceosoma – pre-mRNA interactions is simple based on mechanism of
complementary binds. Participation of more than 200 proteins in splicing also
confirms this idea. We suggest that recognition RNA by spliceosoma is based on
hydropathy interactions, which provide interactions between pre-mRNA and snRNA as well as between pre-mRNA and snRNP.
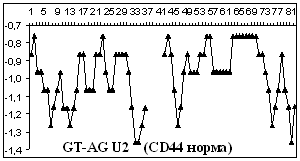
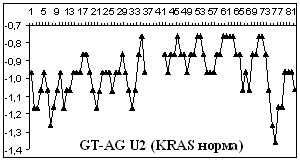
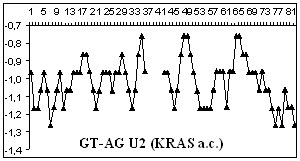
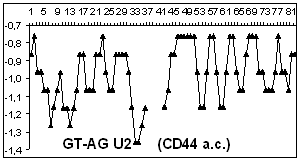
Figure
2 depicts hydropathy profiles of splice sites of three genes responsible for
gastrointestinal tract cancer development (CD44, KRAS and DLEC1). We determine
that splice sites can essentially differ within one gene and even concern to
different types of introns.
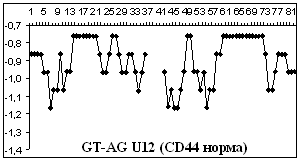
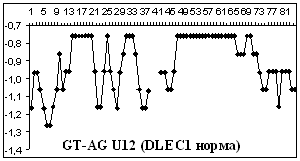
Figure
2. Hydropathy profile of the splice sites of U2 and U12- dependent introns for
CD44, DLEC1 and KRAS genes; x-axis – the numbers of nucleotides, y-axis –
hydropathy values, à.ñ. – alternative variant.
As it is seen from the
Figure 2 advantage
of a choice of 3' splice site remains at isoforms, that code normally
functioning protein, for gene CD44 as well as for gene KRAS.
The character of splicing is control by proteins which
interact with pre-mRNA in ends of introns or exon-intron boundary sites. Such
proteins can block excision of some introns at the same time activating an
excision others one. Pre-mRNA forms tertiary structure to cooperate with other
components of spliceosoma. The splicing regulating proteins could exclude
catalysis of ligation of one pair of exons and will promote effectively
ligation of other one. Directional regulation of splicing ways could be used in
molecular medicine.
References:
1. Nott, A., Meislin S.H., Moore M.J. (2003) A quantitative analysis of
intron effect on mammalian gene expression. RNA. 9, 607-617.
2. Guckian K.M., Schweitzer B.A., Ren
R., X.-F., Sheils C.J., Tahmassebi
D.C., Kool E.T. (2000) Factors conributing
to aromatic stacking in water: evaluation in context of DNA. J. Am. Chem. Soc.,
122, 2213-2222.
3. Chen J., Zhan W., He Y., Peng
J., Wang J., Cai Sh., Ma J. (2004)
Expression of heparanase
gene, CD44v6, MMP-7 and nm23 protein and their relationship with the
invasion and metastasis of gastric carcinomas. World
J Gastroenterol, 15, 776-782.
4. Liu Y., Yan P., Li J., Jia
J. (2005) Expression and significance of CD44s, CD44v6, and nm23 mRNA in human
cancer. World J Gastroenterol,
14, 6601-6606.
5. Cheng Ch., Sharp Ph. (2006) Regulation of CD44
alternative splicing by SRm160 and its potential role in tumor cell invasion.
Molecular and cellular biology, 26, 362-370.
6. Aqeilan R., Kuroki T., Pekarsky
Y., Albagha O., Trapasso
F., Baffa R., Huebner K., Edmonds P., Croce C. (2004) Loss of WWOX Expression in
Gastric Carcinoma. Clinical Cancer Research, 10, 3053-3058.
7. Ishii H., Mimori K., Inageta
T., Murakumo Y., Vecchione
A., Mori M., Furukawa Y. (2005) Components
of DNA Damage Checkpoint Pathway Regulate UV Exposure–Dependent Alterations of Gene
Expression of FHIT and WWOX at Chromosome Fragile Sites. Molecular Cancer Research,
3, 130-138.