PhD student Boldina G.1,
prof. Ivashchenko A.1, prof. Régnier M.2
1 – al-Farabi
Kazakh National University, Almaty,
2 – INRIA,
IDENTIFICATION
REGIONS, WHICH ARE ESSENTIAL FOR SPLICING, ON THE BASE OF HYDROPATHY PROFILES
Pre-mRNA splicing is a nuclear process conserved across eukaryotes.
The spliceosome recognizes conserved sequences at
the exon–intron boundaries.
There are at two classes of pre-mRNA introns, based on the splicing machineries that catalyze
the reaction: U2 and U12 snRNP-dependent introns. Most human introns,
around 99.66% /1/, are likely to be U2- type introns.
The U2-type introns have highly degenerate sequence
motifs. It is still largely unknown how degenerate sequences at the
U2 splice sites are recognized by spliceosome.
In order to find out
regions with conservative properties, namely hydropathy,
which may be recognized by spliceosome and to
distinguish U2 and U12-types of introns, we defined hydropathy
profiles.
Methods
In order to define a general hydropathy profile we built a set
of 313 introns and a set of 385 exons
from genes of 21st
and 22nd chromosomes
contained 1 to 3 introns from GenBank
(http://www.ncbi.nlm.nih.gov).
The flanking sequences (30 nt within the exon and 30 nt
within the intron) were extracted at
both exon–intron junctions,
at 5'ss and 3'ss boundaries. We determined the background hydropathy
value E that is -0.996 for exons and -1.01 for introns.
The corresponding variances are VE = 0.0687
and VI = 0.0690. Regions whose hydropathy
differs from the background value are expected to be essential for recognition
by spliceosome. . At the hydropathy
evaluating, the procedure is given below, the hydropathy
coefficients provided by Guckian et al. in 2000 /2/ were associated to each base i. Given set of splice sites, one
computes an average hydropathy value for each
position as follows. For each base, its number of occurrences at a given
position in the set is multiplied by its hydropathy
coefficient. Summing over all the bases yields the average hydropathy
value. We computed P-value with help
of the large deviation formula /3/ for positions
at the splice sites which deviate from approximately normal distribution. In order to define hydropathy
profiles for splice sites of two subgroups of U2 and U12-type introns we built four sets of 100 introns with confirmed splice sites extracted from SpliceRack database (http://katahdin.cshl.edu:9331/SpliceRack/).
Two sets are associated to human U2-type introns,
with GT–AG and GC–AG termini, and two sets of U12-type introns,
with GT–AG and AT–AC termini correspondingly. For each intron
we extracted 8 nt within the exon and 30 nt
within the intron..
Results
Our method attempts to point out regions which have conservative
properties, namely hydropathy, from a variable
background. Hydropathy profile of genes of 21st and 22nd chromosomes contained
1 to 3 introns is illustrated in Figure 1. For all
pictures the numbers of nucleotides are marked on the x-axis and hydropathy values are indicated by the scale on y-axis.
The termini of the introns are marked in red. Average
values of background hydropathy are marked by red
line. Limits of 99.9% confidence intervals are given by blue dotted lines.
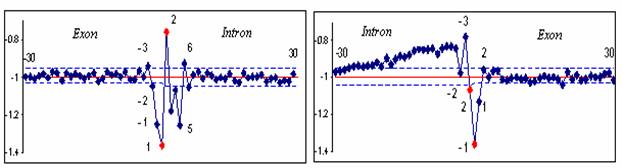
Figure 1. Distinguishing biochemically
conservative regions from background values
The
positions of nucleotides are marked on the x-axis and hydropathy values are indicated by the scale on y-axis.
At positions -30 to -3 within the introns and
+8 to +30 within the exons at the 5’ss and at positions +2 to +30 within the exons at the 3’ss, deviations from the average are
consistent with an approximately normal distribution of hydropathy
values. Slow decay at positions
Regions at the positions -2 to +6 at the 5’ss and -26 to +1 at the 3’ss
deviate from the background hydropathy with
significant P-values. General hydropathy profile of splice cites of genes with 1- 3 introns from 21st
and 22nd chromosomes resembles to the U2-type introns hydropathy profile
(Figure 2), because of the low proportion of U12-type introns
that does not exceed 0,34% /1/.
In
Figures 2 and 3, the hydropathy profiles of U2 and
U12- introns with different termini are depicted.
U2-type
introns
100 splice sites of
U2-type introns with GT-AG as well as with GC-AG
termini extracted
from SpliceRack are considered
separately in order to compare their hydropathy
profiles.
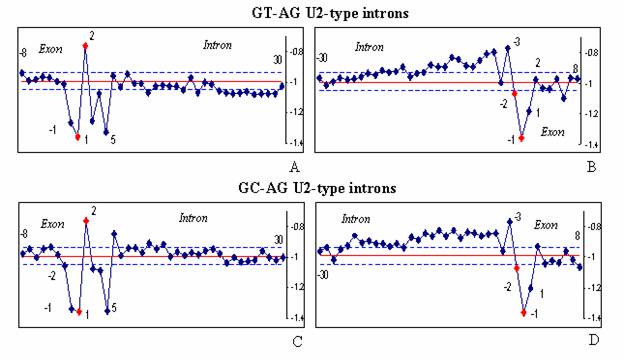
Figure 2. The hydropathy profiles
of the U2-type introns. The hydropathy profiles of two subtypes U2-type introns with GT-AG (A-B) and GC-AG (C-D) termini are shown.
The hydropathy profiles of GT–AG and GC–AG
subtypes are quite similar (Figure 2), Indeed, nucleotide consensus at the 5’ss of U2-type introns mainly contain quite hydrophilic purines when termini are either GT-AG (Figure
U12-type introns.
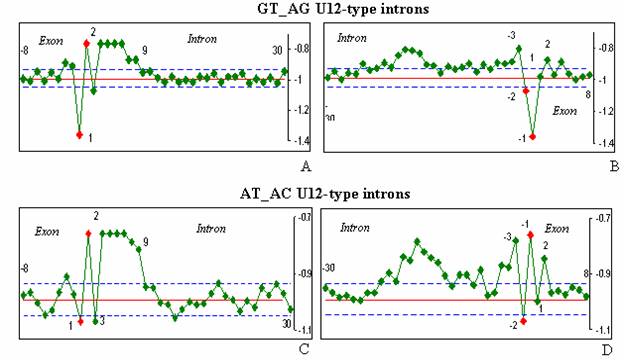
Figure 3. The hydropathy profiles of the U12-type introns.
The hydropathy profiles of two subtypes U12-type introns with GT-AG (A-B) and AT-AC (C-D) termini are shown.
The 5'ss of U12-type introns (Figure
The 3'ss profile for U12-type GT-AG
is different from the U2-type GT-AG (Figures 2B, D and 3B, D). U12-type introns lack obvious PPT at the 3’ss and the BPS lies close
to the
Conclusion
We showed similarity of hydropathy profiles
inside intron types. On the one hand, GT–AG and GC–AG
introns belonging to U2-type have resembling hydropathy profiles as well as AT–AC and GT–AG introns belonging to U12-type. On the other hand, hydropathy profiles of U2 and U12-types GT–AG introns are completely different. Our analysis should be a
step forward for a general understanding of recognition of regions, which are
essential for splicing, by spliceosome and for a
distinction of U2 and U12-types of introns.
References:
1.
Levine, A. and Durbin, R. (2001) A
computational scan for u12-dependent introns in the human genome sequence Nucleic Acids Res,. 29, 4006–4013.
2.
Guckian
K.M., Schweitzer B.A., Ren R., X.-F., Sheils C.J., Tahmassebi D.C., Kool E.T. (2000) Factors conributing
to aromatic stacking in water: evaluation in context of DNA J. Am. Chem. Soc., 122, 2213-2222.
3.
Régnier M., Vandenbogaert
M. (2006) Comparison of statistical significance criteria. Journal of
Bioinformatics and Computational Biology, 4, 537–551.