Halauko Yu.S., Ivashkevich O.À., Matulis Vadim E.
Belarusian State University
DFT Investigation
and QSAR in Series of 5-Nitroimidazoles
Diseases
caused by different pathogenic microorganisms are very widespread and bring a
lot of trouble to mankind. Discovery of natural and development of synthetic
antibiotics in the middle of XX century have led to the illusion about complete
victory in the battle with infectious disorders. Temporary decrease of
pharmaceutical companies` interest to these substances, development of
bacterial resistance and appearance of new diseases have pointed out new
challenges for researches [1]. Introduction of nitroheterocyclic drugs in the
middle of XX century have commemorated a new era in therapy of diseases, caused
by protozoa and some bacteria. 5-Nitroimidazole derivatives represent an
important group of effective antimicrobials which are used as therapeutic
agents against a variety of anaerobic bacterial and protozoal infectious
disorders. These compounds are widely used in medical practice from 1960-s till
nowadays [2]. Metronidazole was the first substance approved for clinical use,
later a few effective analogs of metronidazole were developed and synthesized
[3]. All these substances have nitro group in position “5” of imidazole ring.
Changes in structure leading to changes in physico-chemical properties and also
in biological activity are mainly connected with substituents modification in
position “1” and “2” of imidazole ring. Nitro group causes a high one-electron
reduction potential connected with mechanism of nitroimidazoles action: in
hypoxic cells and anaerobic organisms the reduction of nitro group occurs,
leading to active intermediates, which interact with important biomolecules
(such as DNA), causing death of the cell [2].
QSAR models development is an
important stage of modern drug design process. Analysis of model obtained
allows to recognize clearly essential structural features, responsible for
specific type of activity for further recommendations on lead compounds optimization.
Undoubtedly, that although
considerable proportion of drugs exhibit their activity not as a result of
direct chemical transformations, but as a result of intermolecular interaction
with definite target, this interaction can be described in scientific terms and
can be treated as system perturbation (early stage of chemical reaction). One
of possible ways to achieve this goal is the analysis of substances reactivity
within DFT framework. Hence, molecules biological activity can be characterized
through such descriptors as orbital energies, electrostatic potentials, atomic
charges, hardness and softness and others [4].
Eight different 5-nitroimidazoles
were investigated. All these compounds were used as drugs (some are in use
nowadays). Considered nitroimidazoles were tested against protozoa in test with
clear quantitative endpoint [5] chosen for further modelling. We assume for
convenience the activity of metronidazole in the set of compounds equal to 1,
for other substances activities expressed by relative values (BAr). All
quantum chemical calculations were performed using GAUSSIAN 03W package. Geometric
parameters were calculated by the gradient technique using 6-31G* basis set. To verify that stationary point is true minimum,
force constants and vibrational frequencies were determined (the absence of
imaginary values). 6-311+G** Basis set
have been employed for subsequent energies and other descriptors
calculations. Solvation was treated
using PCM (Polarizable Continuum Model) with standard parameters for water. The
methodology of descriptors calculation, including hydration free energy (Eh), ionization
energy (IE), electron affinity (EA), electronegativity (c), total
hardness (h), electrophilicity index (w) and others, was described
in [6] in details. The names,
biological activities and some of quantum chemical descriptors for investigated
compounds are given in Table.
Table. Nitroimidazole derivatives
under investigation: biological activity and descriptors
|
compound |
descriptor |
|||||||||
|
nonproprietary |
systematic name |
BAr [5] |
IE, |
EA, |
Eh, |
m, |
mi, |
c, |
h, |
w, |
|
metronidazole |
1-(2-hydroxyethyl)-2-methyl-5-nitroimidazole |
1 |
9.32 |
-0.95 |
-5.57 |
3.60 |
0.98 |
4.19 |
5.14 |
1.71 |
|
nimorazole |
1-[2-(4-morpholyl)-ethyl]-5-nitroimidazole |
2.22 |
8.60 |
-0.99 |
-4.01 |
3.65 |
1.26 |
3.81 |
4.80 |
1.51 |
|
ornidazole |
1-(3-chloro-2-hydroxypropyl)-2-methyl-5-nitroimidazole |
1.88 |
9.43 |
-1.13 |
-4.06 |
4.07 |
1.31 |
4.15 |
5.28 |
1.63 |
|
panidazole |
1-[2-(4-pyridyl)-ethyl]-2-methyl-5-nitroimidazole |
0.09 |
8.94 |
-1.06 |
-2.34 |
4.36 |
1.49 |
3.94 |
5.00 |
1.55 |
|
ronidazole |
1-methyl-2-(carbamoyloxymethyl)-5-nitroimidazole |
6.01 |
9.29 |
-0.84 |
-7.64 |
4.78 |
1.39 |
4.23 |
5.07 |
1.76 |
|
secnidazole |
1-(2-hydroxypropyl)-2-methyl-5-nitroimidazole |
0.98 |
9.25 |
-0.88 |
-3.49 |
4.92 |
1.56 |
4.19 |
5.07 |
1.73 |
|
tinidazole |
1-(2-(ethylsulphonyl)-ethyl)-2-methyl-5-nitroimidazole |
1.41 |
9.40 |
-1.12 |
-5.40 |
7.89 |
2.61 |
4.14 |
5.26 |
1.63 |
|
flunidazole |
1-(2-hydroxyethyl)-2-(4-fluorophenyl)-5-nitroimidazole |
4.98 |
8.64 |
-1.24 |
-7.05 |
3.29 |
1.01 |
3.70 |
4.94 |
1.39 |
QSAR model was developed using
multiple linear regression, choosing best model by the criteria of R2 maximization and
minimization of mean standard error. Analysis demonstrates, that the best model
for compounds under consideration includes four descriptors, namely, hydration
energies, dipole moments (m), its`
induction during solvation (mi) and electrophilicity indices.
Act(rel) = -18,46 – 1,84Eh – 8,78m + 23,6mi + 10,8w (1)
R2 = 0,925 F = 9,22 MSE
= 0,76 N
= 8
Figure represents the relationship
between experimental antiprotozoal activity and the prediction 91.
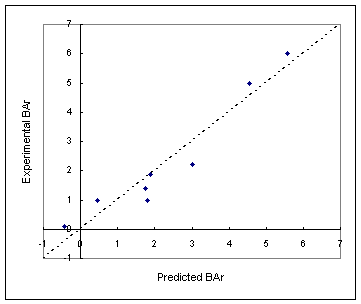
Figure. Experimental versus predicted activities of nitroimidazoles
The
influence of chosen descriptors on biocide activity of nitroimidazoles needs
for some clarification. Hydration free energy calculated by means of DFT can be
used as a measure of substance partition between two phases. Hence, this
descriptor represents membrane transport under conditions close to quasi-static
and can serve as an alternative to widely used empirical octanol/water
partition coefficient logarithm (ClogP).
One can conclude from negative coefficient attached to this descriptor in model
equation, that transport of nitroimidazoles to biological target improves with
hydrophilicity or decreasing of hydration energy. In much the same manner next
descriptor, dipole moment, can be referred to pharmacokinetic phase of drug
action. Activity of compounds decreases with this parameter increase, what can
be related to difficulties in biological barriers penetration. Increase in
activity with electrophilicity index increase can be easily explained,
considering electron transfer process takes place in pharmacodynamic phase of
nitroimidazoles action. The model developed was used to estimate the activity
of a substance with a close activity spectrum – nitazolum (2-acetylamino-5-nitrothiazole).
According to [7] the ratio of nitazolum to metronidazole activity equals 12,
whereas predicted by our model value is 18, which can be judged as good
estimation, regarding change of heterocycle and substituents nature.
A
physically interpretable QSAR model, based on quantum chemical descriptors, for
nitroimidazoles antiprotozoal activity was obtained. We introduced
DFT-calculated hydration free energy as a descriptor, that can be a measure of pharmacokinetic
properties of compounds and serve as alternative to applying empirical models.
It`s potentialities and applicability limits are in need of subsequent
specification. The model obtained can be useful for development of new drugs
carrying nitroazole moiety – potentially with antibacterial or antiprotozoal
action.
1.
Ni, H.; Wendoloski, J. Ann. Rep. Comp. Chem. 2006, 2, 279.
2.
Raether, W.; Haenel, H. Parasitol. Res. 2003, 90,
S19.
3.
Padeyskaya, Å. N. Infect. Antimicrob. Ther. 2000,
2, 110.
4.
Parr, R. G.; Yang, W. Density Functional Theory of Atoms and Molecules; Oxford University
Press: New York, 1989.
5.
Boreham, P. F. L.; Phillips, R. E.; Shepherd, R. W. J. Antimicrob. Chemother. 1985, 16, 589.
6.
Halauko Yu. S., Ivashkevich O. A., Matulis Vadim E. Doklady of the National Academy of Sciences
of Belarus. 2008, 52, 79.
7.
Blatun, L. A.; Lyapunov, N. A.; Kalinichenko, N. F.;
Osolodchenko, T. P.; Svetukhin, A. M.; Pavlova, M. V.; Grishina, I. A.;
Goncharova, Z. G. Antibiot. Khimioter.
1998, 43, 20.