Bulgakova O.V., Ondybaeva
S.B., Kusainova A., and Bersimbay R.I.
L.N.Gumilyov Eurasian
National University
The role of polymorphic genes - GSTM1 and GSTT1 in the pathogenesis of
lung cancer
The
genes that we have studied encode two different forms of glutathione-S-transferases - T1 and M1. These enzymes participate in the second
phase of biotransformation of xenobiotics. Today we know that there are allelic
variants of GSTT1 and GSTM1 genes, and so-called null alleles, which appear
having no protein products at phenotype level. Since these enzymes are the
essential components of the detoxification system, the homozygosis on null
alleles of one or another gene may be associated with high vulnerability of the
organism to the harmful effects and as a consequence it is also connected with
an increased risk of lung cancer. While assessing the results we have, it is
appropriate to recall that the deletion forms of ÑSÒÒ1 and GSTM1 genes, leading
to a lack of appropriate enzymes are considered to be risk factors for breast
cancer, esophageal cancer, cervical cancer, bladder cancer, non-polyposis colon
cancer and lung cancer (LC).
Research
material was collected in the city of Ust- Kamenogorsk (East Kazakhstan) in
2012 -2013 at the Regional Oncology Center. Blood sampling was carried out
according to international rules based on "informed consent" of
volunteers.
DNA
samples were extracted from leukocytic fractions of dark-red blood by a
standard method of phenol and chloroform elution.
Genotyping
of xenobiotics detoxification GSTM1 and GSTT1 genes was carried out by
multiplex polymerase chain reaction method. A mixture for amplification with a
volume of 20 mcl included 30-50 ng of genomic DNA samples, 15 pmol of each
primer (for GSTM1: s 5'-GAA CTC CCT GAA AAG CTA AAG C-3', as 5'-GTT GGG CTC AAA
TAT ACG GTG G-3'; for GSTT1: s 5'-TTC CTT ACT GGT CCT CAC ATC TC-3', as 5'-TCA
CCG GAT CAT GGC CAG CA-3'); 10 mM dNTP, 2 mcl 10õPCR of buffer solution (10 mM
KCL, 100 mM Tris HCl, pH 9.0) and 0.5 units of DNA-polymerase (Sigma, USA).
Conditions
for PCR amplification of GST genes were the following: the initial stage 94o
C, 5 min., then followed by 35 cycles of block: 94 o C, 2 min., 59o
C, 1 min. and 72 o C, 1 min.; and the final stage of elongation – 72o
C, 10 min.
The
amplified products have been analyzed within 2% agarose gel in front of
ethydium bromide and imaging of fragments within UV-light.
The
presence or absence of bands corresponding to the size of 480 b.p. (GSTT1 gene) and 215 b.p. (GSTM1 gene)
allowed to estimate the presence or absence of deletion polymorphisms.
Polymorphic
alleles of GST genes, having extensive deletions, did not amplified, and
therefore there were no bands with the size of 215 b.p. and 480 b.p. as it was
mentioned above.
Thus, the genotyping method helped to distinguish homozygotic deletion
of GST gene (null genotype, 0/0) from a heterozygote and a normal homozygote
(plus Genotype +/+ and +/0).
The
results of genotyping GSTM1 and GSTT1 genes are represented in the form of
electrophoregrams (Figure 1.2). According to the results of this study among
the patients with LC there is a higher incidence of null genotypes of GSTT1-
51.4% and GSTM1 - 59.4%, than in the group under control - 15 and 43 %
(p<0.01).
OR
values indicate that the null genotype is associated with
underlying risk for lung cancer, and it is when the genotype of 0/0 GSTT1 has a
greater risk significance than 0/ 0 GSTM1 (OR = 5.99; Ñ195î/î=1.16-3.32
and OR = l.96; Cl95o/0= 1.49-5.03, respectively).
According to the results of our study the carriers of both deletions of GSTM1
and GSTT1 genes are more subject to risk of lung cancer than the carriers of
the normal genotype GSTT1 (+) and GSTM1 (+).
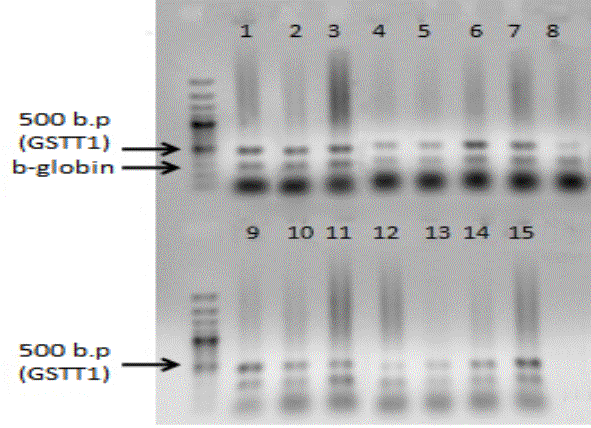
Figure 1 - Examples of
identification 0/0 + genotypes on GSTT1 gene (480 base pair)
215 b. p. (GSTM1) 268 b. p. (b-globin) 480 b. p. (GSTT1) ![]()
![]()
![]()
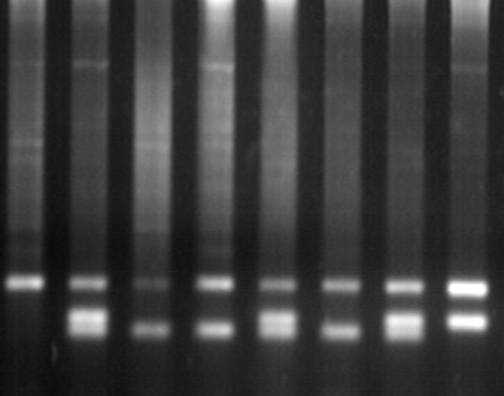
Figure 2 - Examples of
identification 0/0 + genotypes on GSTM1
gene (215 base pair)