Sbasheva L.M., Akimbekov N.Sh.
al-Farabi Kazakh National University,
Almaty, Kazakhstan
Isolation and identification of lactic acid bacteria in milk samples from the South Kazakhstan region
Raw countryside cow milk samples delivered from the South
Kazakhstan region was investigated for the presence of lactic acid bacteria
(LAB). A total of 13 microorganisms were isolated and only 2 of them were
determined as LAB. Morphological, cultural and genetic analysis were performed
to identify the strains. As a result these strains were defined as Lactobacillus
plantarum JJN and Leuconostoc mesenteroides JJN-2. The current
research constitutes the first step in the development of a new dairy product
based on the indigenous LAB strains.
INTRODUCTION
Lactic
acid bacteria (LAB) are widely distributed in nature and occur naturally as
indigenous microflora in raw milk and fermented dairy products. They are
gram-positive bacteria that play an important role in many food processing. The
species of the genera Leuconostoc (Ln.) and Lactobacillus (Lb.)
are included in this group [1]. In addition, they strongly determine the flavor,
texture and frequently, the nutritional value of food and feed products [2-3].
Lactic
acid bacteria perform an essential role in the preservation and production of
wholesome foods. The lactic acid fermentations are generally inexpensive, and
often little or no heat is required in their preparation, making them fuel
efficient as well. Foods fermented with lactic acid play an important role in
feeding the world's population on every continent.
Lactic
acid bacteria perform this essential function in preserving and producing a
wide range of foods: fermented fresh vegetables such as cabbage (sauerkraut,
Korean kimchi); cucumbers (pickles);
fermented cereal yogurt (Nigerian ogi,
Kenyan uji); sourdough bread and
bread-like products made without wheat or rye flours (Indian idli, Philippine puto); fermented milks (yogurts and cheeses); fermented milk-wheat
mixtures (Egyptian kishk, Greek trahanas); protein-rich vegetable
protein meat substitutes (Indonesian tempe);
amino acid/peptide meat-flavored sauces and pastes produced by fermentation of
cereals and legumes (Japanese miso,
Chinese soy sauce); fermented cereal-fish-shrimp mixtures (Philippine balao balao and burong dalag); and fermented meats (e.g., salami).
Lactic
acid bacteria are generally fastidious on artificial media, but they grow
readily in most food substrates and lower the pH rapidly to a point where
competing organisms are no longer able to grow. Leuconostocs and lactic streptococci generally thrive in a lower
pH, about 4-4.5 and some of the lactobacilli and pediococci can grow about pH
3.5, before inhibiting their own growth [4].
The
objective of present study was to isolate and identify the indigenous LAB
species from cows’ raw milk samples from the South
Kazakhstan region in order to use them in the interests of the further
functional product development based on LAB.
MATERIALS AND METHODS
Milk samples
Cows’ raw milk samples were taken from the South
Kazakhstan countryside farm in January 2016. The milk was collected into a
sterile bottle and immediately transported to the
laboratory under proper environmental conditions and in a timely manner. The
samples were kept at 4°C.
Figure 1. The
location of the provided samples: Kegen region Almaty district, South
Kazakhstan
Isolation and
characterization of strains
Milk agar and MRS agar were used for isolation
of LAB. The milk was diluted to 1:100 and 1:1000. 0,1 ml of these
dilutions was inoculated on petri dishes with nutrient medium, and incubated
for 72h at 37°C. After incubation the CFU
morphologically most resembling LAB were selected and streaked on pure agar
plates. Cell morphology was determined by Gram staining and examination under
the microscope. Finally, pure colonies were cultivated and stored on MRS and
Milk agar slants at 4°C.
DNA isolation and sequence analysis
For sequencing analysis bacteria were cultivated in MRS broth at 37°C
for 48 h. Bacterial culture taken from Milk agar was labeled as sample #1 and
culture from MRS agar was labeled as sample #2.
Bacterial genomic DNA was isolated using the kit PureLink® Genomic DNA
Kits (Invitrogene, USA). The DNA concentration in the samples was determined
using the Qubit fluorimeter (Invitrogen, USA) according to the scale of HS
dsDNA.
Molecular-genetic identification of microorganisms was carried out based
on the study of phase sequence of the 16S rRNA gene using universal primers 8F
(5'-AGAGTTTGATCCTGGCTCAG-3') and 806R (5'-GGACTACCAGGGTATCTAAT-3') (Edwards et
al.).
The reaction mixture (30 µl) contained 3 µl of 10x reaction buffer
(Fermentas), 2,5 mM MgCl2, 0.2 mM of each deoxyribonucleotides (dNTP), 10 pmol
of each of the primers, 1 unit of Taq polymerase Maxima Hot Start Taq DNA
Polymerase (Fermentas). PCR was carried out in a thermal cycler Mastercycler
proS (Eppendorf).
The reaction was started by incubation of the mixture at 95 °C for 7
min, followed by 30 cycles consisting of: 95 °C 30 seconds, 55 °C 40 seconds ,
72 °C – 1 min. Final elongation was performed at 72 °C for 10 minutes.
Amplificatory product was separated in 1.2% agarose gel, the bands were stained
with ethidium bromide and visualized using the documentation system gel
INFINITY VX2 (made in VILBER LOURMAT, France). As the electrode buffer used 1õÒÀÅ-buffer. PCR
products were purified using the kit PureLink® PCR Purification Kit
(Invitrogen, USA).
Sequencing of 16S rRNA bacterial gene fragments was performed using
BigDye® Terminator v3.1 Cycle Sequencing Kit according to the manufacturer's
Protocol [BigDye® Terminator v3.1 Cycle Sequencing Kit Protocol Applied
Biosystems, USA]. The sequencing products obtained with BidDye 3.1 terminator
were purified using a reagent for the purification of PCR products PCR Purification
Reagent CleanSweep.
Capillary electrophoresis of sequencing products was performed on an
automatic sequencer 3500 DNA Analyzer (Applide Biosystems, USA).
Sequence results were processed in the program SeqÀ6 (Applide
Biosystems). Search of homologous nucleotide sequences of 16S rRNA genes was
performed using the program BLAST (Basic Local Alignment Search Tool) in the
International database Gene Bank of the National Center for Biotechnology
Information, USA (htpp://www.ncbi. nlm. nih.gov). Phylogenetic analysis was
performed using the software MEGA6. Alignment of nucleotide sequences was
performed using the algorithm ClustalW. To build phylogenetic trees we used the
method of "Association of neighbors" Neiighbor-Joining (NJ).
RESULTS
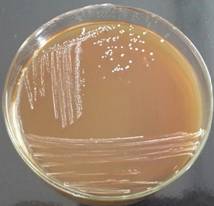
Morphological analysis of strains
Pure
colonies obtained on Milk agar were small (< 1mm), round shaped, and had
creamy-white colour. They were Gram-positive, elongated, paired cocci. Colonies
grown on MRS agar were also small (1-2 mm), round shaped and had bright white
colour. They were Gram-positive and had elongated, rod shaped cell morphology
(Fig. 2-3).
|
|
|
|
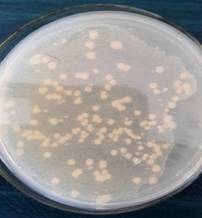
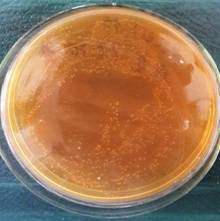
CFU
on Milk Agar (1:1000) |
CFU on MRS Agar (1:1000) |
|
|
|
|
Colonies
of Ln. mesenteroides |
Colonies of Lb. plantarum |
Figure
2. Cultural characteristics of LAB
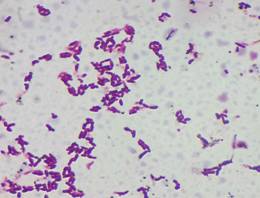
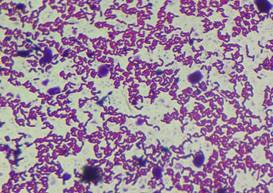
|
Ln.
mesenteroides |
Lb. plantaum |
Figure 3. Morphological characteristics of LAB
DNA sequencing
DNA concentrations in the samples were: sample #
1 – 12,8 ɲg/µI; # 2 – 2KG – 33,8 ɲg/µI.
After amplification with primers for the 16S rRNA was the PCR product with a size of about 650 bp was obtained.
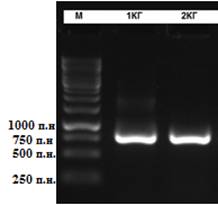
Figure 4 – PCR product obtained after amplification with universal
primers
Note: 1,2 – bacterial samples; M – molecular mass marker O'GeneRuler 1 kb
DNA Ladder.
The concentration of PCR product after purification was as follows:
sample 1 – 62.6 per ɲg/µI and in the sample 2 – 70,26 ɲg/µI.
After conducting capillary electrophoresis on the 3500 instrument DNA
Analyzer data were processed in the program SeqÀ6. As the result the following nucleotide
sequences were obtained:
The nucleotide sequence of the strain 1:
GAGTTGAGCTCCGGGCTTTCACATCAGACTTAATAAACCGTCTGCGCTCGCTTTACGCCCAATAAATCCGGATAACGCTCGGGACATACGTATTACCGCGGCTGCTGGCACGTATTTAGCCGTCCCTTTCTGGTATGGTACCGTCAAACTAAAATCATTTCCTATTCTAGCTGTTCTTCCCATACAACAGTGCTTTACGACCCGAAAGCCTTCATCACACACGCGGCGTTGCTCCATCAGGCTTTCGCCCATTGTGGAAGATTCCCTACTGCAGCCTCCCGTAGGAGTTTGGGCCGTGTCTCAGTCCCAATGTGGCCGATCAGTCTCTCAACTCGGCTATGCATCATTGTCTTGGTAGGCCTTTACCCCACCAACTAACTAATGCACCGCGGATCCATCTCTAGGTGACGCCGAAACGCCTTTTAACTTTGTGTCATGCGACACTAAGTTTTATTCGGTATTAGCATCTGTTTCCAAATGTTATCCCCAGCCTTGAGGCAGGTTGTCCACGTGTTACTCACCCGTTCGCCACTCACTTGAAAGGTGCAAGCACCTTTCGCTGTGCGTTCGACTTGCAT
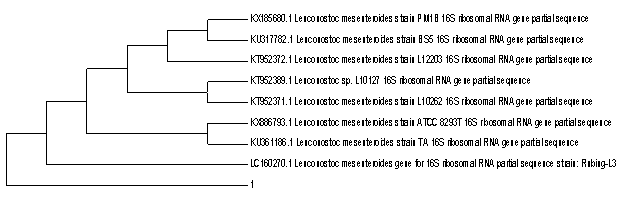
Search of homologous nucleotide sequences of 16S rRNA genes by the program BLAST (Basic Local Alignment Search Tool) in the International database Gene Bank of the National center for biotechnology information, USA (htpp://www.ncbi. nlm. nih.gov) showed the identity of the studied strain to the species Leuconostoc mesenteroides (99% homology).
Nucleotide sequence of the strain 2:
GGGGATAACACCTGGAAACAGATGCTAATACCGCATAACAACTTGGACCGCATGGTCCGAGTTTGAAAGATGGCTTCGGCTATCACTTTTGGATGGTCCCGCGGCGTATTAGCTAGATGGTGGGGTAACGGCTCACCATGGCAATGATACGTAGCCGACCTGAGAGGGTAATCGGCCACATTGGGACTGAGACACGGCCCAAACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCACAATGGACGAAAGTCTGATGGAGCAACGCCGCGTGAGTGAAGAAGGGTTTCGGCTCGTAAAACTCTGTTGTTAAAGAAGAACATATCTGAGAGTAACTGTTCAGGTATTGACGGTATTTAACCAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTGGCAAGCGTTGTCCGGATTTATTGGGCGTAAAGCGAGCGCAGGCGGTTTTTTAAGTCTGATGTGAAAGCCTTCGGCTCAACCGAAGAAGTGCATCGGAAACTGGGAAACTTGAGTGCAGAAGAGGACAGTGGAACTCCATGTGTAGCGGTGAAATGCGTAGATATATGGAAGAACACCAGTGGCGAAGGCGGCTGTCTGGTCTGTAACTGACGCTGAGGCTCGAAAGTATGGGTAGCAAACAGGATTAGATACCCTGGTAGTC
Search of homologous nucleotide sequences of the genes 16S rRNA by using the program BLAST (Basic Local Alignment Search Tool) in the International database Gene Bank of the National center for biotechnology information, USA (htpp://www.ncbi. nlm. nih.gov) showed the identity of the studied strain to the species Lactobacillus plantarum (100% homology).As can be seen from the phylogenetic tree, nearest-neighbor strain is Lactobacillus plantarum HUMBO7393.

The leuconostocs are included primarily as flavor producers, in
combination with strains of mesophilic acid producers such as Lactococcus.
Citrate-fermenting strains of leuconostocs impart buttery flavor via diacetyl
formation. The primary fermentation products from lactose are d(–)-lactate,
ethanol, and CO2. Acetic acid can also be produced. Some
leuconostocs produce dextrans and can also be directed toward the production of
oligosaccharides, which may have textural (hydrocolloid) and functional
(prebiotic) application in dairy foods. Leuconostocs are not notably
proteolytic or lipolytic. Some strains of Leuconostoc produce an antilisterial
bacteriocin [6]. L.plantarum is considered a probiotic because it secretes
antimicrobial compounds, such as bacteriocin, that inhibit pathogenic
gram-positive and gram-negative colonies from forming [7]. The bacterium gives
food certain tastes and flavors depending on the balance between acetate
(volatile) and lactate (nonvolatile) organic acids [8]. Such properties of
L.mesenteroides and L.plantarum, and in addition them being indigenous strains,
can be usefully applied to the development of a new dairy product.
CONCLUSION
In this study, traditional culture method and gene analysis were applied
to analyze the composition of unfermented cow milk from the South Kazakhstan. A
total of 13 bacteria were isolated and 2 of them were identified and classified
into LAB. Lactobacillus plantarum JJN and Leuconostoc mesenteroides JJN-2 were considered as the predominated
LAB species among all samples under the condition of cultivating in MRS culture
medium. This research contributes to an understanding of the composition of LAB
in raw milk of the South Kazakhstan, which could provide important data and
strain resources for further study involved in strain selection and starter
culture design for preparing functional drinks and foods.
REFERENCES
1. P. Azadnia,
A. Khalegh Babaki,
M. Shah Ahmad Ghasemi,
M. Karimi Jashni,
M.H. Zamani
and N. Taarof. Isolation and Identification of Leuconostocs
from Traditional Yoghurt in Tribes of Kazerun // Journal of animal and
veterinary advances. - 2011. - Vol. 10
P. 552-554.
2. Lee B.H. Bacteria based processes
and products // Fundamentals of food biotechnology, Lee B.H. (ed.) John Wiley
and Sons, New York, 1996. P. 219-290.
3. Lilia Tserovska*, Stanka Stefanova and Tanya Yordanova. Identification of lactic acid bacteria isolated from katyk, goat's milk and cheese // Journal of culture collections. - 2002. - Vol. 3. P. 48-52.
4. https://www.ncbi.nlm.nih.gov/books/NBK234703/
5. Edwards U., Rogall T., Blocker H., Emde, M., Bottger, E. C. Isolation
and direct complete nucleotide determination of entire genes: characterization of
a gene coding for 16S ribosomal RNA // Nucleic. Acids Res. – 1989. – Vol. 17.
P. 7843-7853.
6. R. Holland S.-Q. Liu. Lactic acid bacteria | Leuconostoc spp. // Encyclopedia of Dairy Sciences (Second Edition) – 2011.- P. 138-142.